

Journal of Clinical and Biomedical Sciences
DOI: 10.58739/jcbs/v15i4.25.112
Year: 2025, Volume: 15, Issue: 4, Pages: 257-267
Original Article
Seyedeh Maryam Mousavi1*, Banafsheh Sadat Torabi2, Amin Hashemi3, Somayeh Mohammadi4, Masoud Faraji5, Mohsen Sadeghi6
1Department of Biology, Faculty of Sciences, Shahid Chamran University of Ahvaz, Ahvaz, Iran.
2Department of Biology, Faculty of Sciences, Islamic Azad University, Damghan, Iran.
3Department of Microbiology, Faculty of Medicine, Shahid Sadoughi University of Medical Sciences and Health Services, Yazd, Iran.
4Department of Microbiology, Faculty of Basic Sciences, Islamic Azad University, Lahijan, Iran.
5Department of Microbiology, Faculty of Sciences, Islamic Azad University, Karaj, Iran.
6Department of Microbiology, Faculty of Advanced Sciences and Technology, Islamic Azad University, Tehran Medical Branch, Tehran, Iran.
*Corresponding Author
Email: [email protected]
Received Date:05 March 2025, Accepted Date:30 May 2025, Published Date:21 December 2025
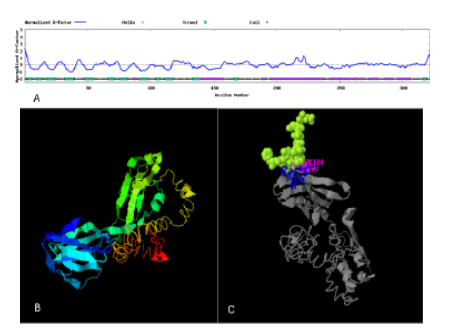
Helicobacter pylori is a pathogen that has been consistently linked to gastrointestinal cancer, particularly gastric cancer. Reports indicate that this pathogen is resistant to antibiotic treatments, necessitating the development of new treatment strategies. Here, to design a vaccine against H. pylori immunoinformatic tools were used. For this purpose, five virulence proteins including Flagellin B (FlaB), Flagellin A (FlaA), Urease subunit beta (UreB), CagA and Vacuolating cytotoxin autotransporter (VacA) were selected. Antigenicity and allergenicity were evaluated, and the epitopes with the highest scores were chosen. Linkers were used to connect the epitopes and an extracellular domain of CTLA-4 was positioned on the N-terminal. Biochemical features were predicted with the ProtParam server, and the second structure was predicted with the Prabi server, while the third structure was predicted with the Robetta, Alphafold and I-TASSER servers. VaxiJen and AllerTOP servers were used to evaluate the vaccine's antigenicity and allergenicity, respectively. After confirming the structure of the designed vaccine, molecular docking was performed with the TLR5 molecule using AutoDock Vina software. The SnapGene tool was utilized to in silico cloning of the vaccine in pET-3a vector. To evaluate the efficacy of the recombinant vaccine, the multi-epitope gene of H. pylori was cloned into the pCDNA3.1 vector, and its expression was analyzed in the spleen tissue of BALB/c mice using RT-qPCR. It has been shown in the results that the vaccine designed can bind to the TLR5 molecule on the surface of immune cells. Despite being an antigen, this vaccine didn't have any allergenic properties. Immunoinformatic is a promising tool for designing various drugs and vaccines. The results demonstrated that the expression levels of TNF-α and IFN-γ were significantly increased in the vaccine-treated group compared to the control group (P < 0.05). The elevated expression of pro-inflammatory cytokines indicates that the designed DNA vaccine successfully induced a cellular immune response against H. pylori.
Keywords: H. pylori; Vaccine design; CTLA-4; Immunoinformatic; Gastric cancer
1. Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. The Lancet. 1983; 321 (8336). Available from: https://doi.org/10.1016/s0140-6736(83)92719-8
2. Sit WY, Chen YA, Chen YL, Lai CH, Wang WC. Cellular evasion strategies of Helicobacter pylori in regulating its intracellular fate. Seminars in Cell & Developmental Biology. 2020; 101 Available from: https://doi.org/10.1016/j.semcdb.2020.01.007
3. Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, et al. Chronic inflammation in the etiology of disease across the life span. Nature Medicine. 2019; 25 (12). Available from: https://doi.org/10.1038/s41591-019-0675-0
4. Ražuka‐Ebela, D, Giupponi B, Franceschi F. Helicobacter pylori and extragastric diseases. Helicobacter. 2018; 23 (S1). Available from: https://doi.org/10.1111/hel.12520
5. Torok AM, Bouton AH, Goldberg JB. Helicobacter pyloriInduces Interleukin-8 Secretion by Toll-Like Receptor 2- and Toll-Like Receptor 5-Dependent and -Independent Pathways. Infection and Immunity. 2005; 73 (3). Available from: https://doi.org/10.1128/iai.73.3.1523-1531.2005
6. Junaid M, Shah M, Khan A, Li CD, Khan MT, Kaushik AC, et al. Structural-dynamic insights into the H. pylori cytotoxin-associated gene A (CagA) and its abrogation to interact with the tumor suppressor protein ASPP2 using decoy peptides. Journal of Biomolecular Structure and Dynamics. 2019; 37 (15). Available from: https://doi.org/10.1080/07391102.2018.1537895
7. Marques AT, Vítor JMB, Santos A, Oleastro M, Vale FF. Trends in Helicobacter pylori resistance to clarithromycin: from phenotypic to genomic approaches. Microbial Genomics. 2020; 6 (3). Available from: https://doi.org/10.1099/mgen.0.000344
8. Suzuki S, Esaki M, Kusano C, Ikehara H, Gotoda T. Development of Helicobacter pylori treatment: How do we manage antimicrobial resistance?. World Journal of Gastroenterology. 2019; 25 (16). Available from: https://doi.org/10.3748/wjg.v25.i16.1907
9. Ghosh P, Bhakta S, Bhattacharya M, Sharma AR, Sharma G, Lee SS et al. A Novel Multi-Epitopic Peptide Vaccine Candidate Against Helicobacter pylori: In-Silico Identification, Design, Cloning and Validation Through Molecular Dynamics. International Journal of Peptide Research and Therapeutics. 2021; 27 (2). Available from: https://doi.org/10.1007/s10989-020-10157-w
10. Chen J, Lin L, Li N, She F. Enhancement of Helicobacter pylori outer inflammatory protein DNA vaccine efficacy by co‐delivery of interleukin‐2 and B subunit heat‐labile toxin gene encoded plasmids. Microbiology and Immunology. 2012; 56 (2). Available from: https://doi.org/10.1111/j.1348-0421.2011.00409.x
11. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD et al. Protein Identification and Analysis Tools on the ExPASy Server. The Proteomics Protocols Handbook. 2005; Available from: https://doi.org/10.1385/1-59259-890-0:571
12. Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. "Protein Engineering, Design and Selection". 1994; 7 (2). Available from: https://doi.org/10.1093/protein/7.2.157
13. Razavi A, Bagheri N, Azadegan-Dehkordi F, Shirzad M, Rahimian G, Rafieian-Kopaei M, et al. Comparative Immune Response in Children and Adults withH. pyloriInfection. Journal of Immunology Research. 2015; 2015 Available from: https://doi.org/10.1155/2015/315957
14. Moyat M, Velin D. Immune responses toHelicobacter pyloriinfection. World Journal of Gastroenterology. 2014; 20 (19). Available from: https://doi.org/10.3748/wjg.v20.i19.5583
15. Menheniott TR, Judd LM, Giraud AS. STAT3: a critical component in the response toHelicobacter pyloriinfection. Cellular Microbiology. 2015; 17 (11). Available from: https://doi.org/10.1111/cmi.12518
16. Jafarzadeh A, Larussa T, Nemati M, Jalapour S. T cell subsets play an important role in the determination of the clinical outcome of Helicobacter pylori infection. Microbial Pathogenesis. 2018; 116 Available from: https://doi.org/10.1016/j.micpath.2018.01.040
17. Algood HMS, Cover TL. Helicobacter pyloriPersistence: an Overview of Interactions betweenH. pyloriand Host Immune Defenses. Clinical Microbiology Reviews. 2006; 19 (4). Available from: https://doi.org/10.1128/cmr.00006-06
18. Larussa T, Leone I, Suraci E, Imeneo M, Luzza F. Helicobacter pyloriand T Helper Cells: Mechanisms of Immune Escape and Tolerance. Journal of Immunology Research. 2015; 2015 Available from: https://doi.org/10.1155/2015/981328
19. Mohammadi M, Nedrud J, Redline R, Lycke N, Czinn SJ. Murine CD4 T-cell response to Helicobacter infection: TH1 cells enhance gastritis and TH2 cells reduce bacterial load. Gastroenterology. 1997; 113 (6). Available from: https://doi.org/10.1016/s0016-5085(97)70004-0
20. Schmitz J, Thiel A, Kühn R, Rajewsky K, Müller W, Assenmacher M, et al. Induction of interleukin 4 (IL-4) expression in T helper (Th) cells is not dependent on IL-4 from non-Th cells.. The Journal of experimental medicine. 1994; 179 (4). Available from: https://doi.org/10.1084/jem.179.4.1349
21. Rivera-Hernandez T, Rhyme MS, Cork AJ, Jones S, Segui-Perez C, Brunner L, et al. Vaccine-Induced Th1-Type Response Protects against Invasive Group A Streptococcus Infection in the Absence of Opsonizing Antibodies. mBio. 2020; 11 (2). Available from: https://doi.org/10.1128/mbio.00122-20
22. Gu H. Role of Flagella in the Pathogenesis of Helicobacter pylori. Current Microbiology. 2017; 74 (7). Available from: https://doi.org/10.1007/s00284-017-1256-4
23. Clyne M, Ocroinin T, Suerbaum S, Josenhans C, Drumm B, et al. Adherence of Isogenic Flagellum-Negative Mutants of Helicobacter pylori and Helicobacter mustelae to Human and Ferret Gastric Epithelial Cells. Infection and Immunity. 2000; 68 (7). Available from: https://doi.org/10.1128/iai.68.7.4335-4339.2000
24. Yan J, Liang SH, MaoYF, Li LW, Li P. Construction of expression systems forflaAandflaBgenes ofHelicobacter pyloriand determination of immunoreactivity and antigenicity of recombinant proteins. World Journal of Gastroenterology. 2003; 9 (10). Available from: https://doi.org/10.3748/wjg.v9.i10.2240
25. Blanchard TG, Czinn SJ. Identification of Helicobacter pylori and the evolution of an efficacious childhood vaccine to protect against gastritis and peptic ulcer disease. Pediatric Research. 2017; 81 (1-2). Available from: https://doi.org/10.1038/pr.2016.199
26. Pan X, Ke H, Niu X, Li S, Lv J, Pan L. Protection Against Helicobacter pylori Infection in BALB/c Mouse Model by Oral Administration of Multivalent Epitope-Based Vaccine of Cholera Toxin B Subunit-HUUC. Frontiers in Immunology. 2018; 9 Available from: https://doi.org/10.3389/fimmu.2018.01003
27. Moyat M, Velin D. Use of VacA as a Vaccine Antigen. Toxins. 2016; 8 (6). Available from: https://doi.org/10.3390/toxins8060181
28. Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003; 125 (6). Available from: https://doi.org/10.1053/j.gastro.2003.08.033
29. Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection.. Gut. 1997; 40 (3). Available from: https://doi.org/10.1136/gut.40.3.297
30. Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nature Reviews Gastroenterology & Hepatology. 2010; 7 (11). Available from: https://doi.org/10.1038/nrgastro.2010.154
31. van Hooren L, Sandin LC, Moskalev I, Ellmark P, Dimberg A, Black P, et al. Local checkpoint inhibition of CTLA‐4 as a monotherapy or in combination with anti‐PD1 prevents the growth of murine bladder cancer. European Journal of Immunology. 2017; 47 (2). Available from: https://doi.org/10.1002/eji.201646583
32. Boyle JS, Brady JL, Lew AM. Enhanced responses to a DNA vaccine encoding a fusion antigen that is directed to sites of immune induction. Nature. 1998; 392 (6674). Available from: https://doi.org/10.1038/32932
33. O'neill LAJ, Golenbock D, Bowie AG. The history of Toll-like receptors — redefining innate immunity. Nature Reviews Immunology. 2013; 13 (6). Available from: https://doi.org/10.1038/nri3446
34. Ishihara S, Rumi MAK, Kadowaki Y, Ortega-Cava CF, Yuki T, Yoshino N, et al. Essential Role of MD-2 in TLR4-Dependent Signaling during Helicobacter pylori-Associated Gastritis. The Journal of Immunology. 2004; 173 (2). Available from: https://doi.org/10.4049/jimmunol.173.2.1406
35. Ru Z, Yu M, Zhu Y, Chen Z, Zhang F, Zhang Z, et al. Immmunoinformatics‐based design of a multi‐epitope vaccine with CTLA‐4 extracellular domain to combat Helicobacter pylori. The FASEB Journal. 2022; 36 (4). Available from: https://doi.org/10.1096/fj.202101538rr
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Published By Sri Devaraj Urs Academy of Higher Education, Kolar, Karnataka
Subscribe now for latest articles and news.