

Journal of Clinical and Biomedical Sciences
DOI: 10.58739/jcbs/v15i4.25.139
Year: 2025, Volume: 15, Issue: 4, Pages: 277- 283
Original Article
Seyedeh Maryam Mousavi1*, Soroush Karimi2, Atefeh Azadi3, Mahsa Boogari4, Hossein Joveini5, Arman Izadian6
1Department of Biology, Faculty of Sciences, Shahid Chamran University of Ahvaz, Ahvaz, Iran.
2Nano Drug Delivery Research Center, Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran.
3Department of Laboratory Sciences, Faculty of Paramedicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
4Department of Molecular Medicine, Pasteur Institute of Iran, Tehran, Iran.
5Department of Laboratory Sciences, Faculty of Medical Sciences, Islamic Azad University, Gorgan Branch, Gorgan, Iran.
6Department of Vector Biology and Control of Diseases, School of Public Health,Tehran University of Medical Sciences, Tehran, Iran.
*Corresponding Author
Email: [email protected]
Received Date:24 March 2025, Accepted Date:29 July 2025, Published Date:31 December 2025
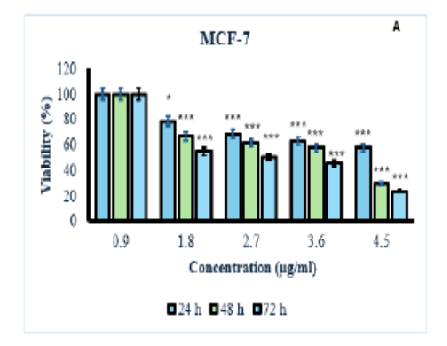
A protein called cytolysin A or ClyA, encoded by certain bacteria species, can cause cytotoxicity. Although the ClyA protein is not typically expressed at detectable levels in most E. coli strains, here it was successfully overproduced and purified by cloning the structural gene into a hns mutant strain. The cytotoxicity of the purified cytolysin was assessed on two MCF-7 cancer cell lines and HDF normal cell line using the MTT assay. Flow cytometry was employed to examine the cytolysin's ability to induce apoptosis in cancer cells. In addition, a Western blot analysis was carried out to evaluate the expression levels of P53, Bcl2, and Bax proteins. The results revealed that cytolysin exhibited dose-dependent and time-dependent toxicity towards cancer cells, while showing minimal toxicity against normal cells, indicating its selective action against cancer cells. Cytolysin had an IC50 value of 3.29 µg/ml against MCF-7 cancer cells and 12.6 µg/ml against HDF normal cells. Flow cytometry results further demonstrated that cytolysin induced apoptosis in cancer cells, evidenced by increased expression of p53 and BCL2, as well as decreased in Bax, in gene and protein levels. These findings underscore the potential of cytolysin as a targeted therapy for cancer, highlighting its selective cytotoxic effect on cancer cells.
Keywords: Cytolysin-A, Breast cancer, Cytotoxicity, Flowcytometry, Western blotting
1. Oscarsson J, Mizunoe Y, Uhlin BE, Haydon DJ. Induction of haemolytic activity in <I>Escherichia coli</I> by the slyA gene product. Molecular Microbiology. 1996; 20 (1). Available from: https://doi.org/10.1111/j.1365-2958.1996.tb02500.x
2. Ludwig A, Tengel C, Bauer S, Bubert A, Benz R, Mollenkopf HJ, <I>et al</I>. SlyA, a regulatory protein from <I>Salmonella typhimurium</I>, induces a haemolytic and pore-forming protein in <I>Escherichia coli</I>. Molecular and General Genetics MGG. 1995; 249 (5). Available from: https://doi.org/10.1007/bf00290573
3. Murase K, Ooka T, Iguchi A, Ogura A, Nakayama K, Asadulghani M, <I>et al</I>. Haemolysin E-and enterohaemolysin-derived haemolytic activity of O55/O157 strains and other <I>Escherichia coli</I> lineages. Microbiology. 2012; 158 (3). Available from: https://doi.org/10.1099/mic.0.054775-0
4. Ludwig A, Von Rhein C, Bauer S, Hüttinger C, Goebel W. Molecular analysis of cytolysin A (ClyA) in pathogenic <I>Escherichia coli</I> strains. Journal of Bacteriology. 2004; 186 (16). Available from: https://doi.org/10.1128/jb.186.16.5311-5320.2004
5. Enow COA, Oscarsson J, Zlatkov N, Westermark M, Duperthuy M, Wai SN, <I>et al</I>. Elevated recombinant clyA gene expression in the uropathogenic <I>Escherichia coli</I> strain 536, a clue to explain pathoadaptive mutations in a subset of extraintestinal <I>E. coli</I> strains. BMC Microbiology. 2014; 14 (1). Available from: https://doi.org/10.1186/s12866-014-0216-4
6. Del Castillo FJ, Leal SC, Moreno F, Castillo Id. The <I>Escherichia coli</I> K‐12 sheA gene encodes a 34‐kDa secreted haemolysin. Molecular Microbiology. 1997; 25 (1). Available from: https://doi.org/10.1046/j.1365-2958.1997.4391813.x
7. Oscarsson J, Westermark M, Löfdahl S, Olsen B, Palmgren H, Mizunoe Y, <I>et al</I>. Characterization of a pore-forming cytotoxin expressed by <I>Salmonella enterica</I> serovars Typhi and Paratyphi A. Infection and Immunity. 2002; 70 (10). Available from: https://doi.org/10.1128/iai.70.10.5759-5769.2002
8. Von Rhein C, Bauer S, Simon V, Ludwig A. Occurrence and characteristics of the cytolysin A gene in <I>Shigella</I> strains and other members of the family Enterobacteriaceae. FEMS Microbiology Letters. 2008; 287 (2). Available from: https://doi.org/10.1111/j.1574-6968.2008.01290.x
9. von Rhein C, Bauer S, Sanjurjo EJL, Benz R, Goebel W, Ludwig A. ClyA cytolysin from <I>Salmonella</I>: distribution within the genus, regulation of expression by SlyA, and pore-forming characteristics. International Journal of Medical Microbiology. 2009; 299 (1). Available from: https://doi.org/10.1016/j.ijmm.2008.06.004
10. Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature. 2009; 459 (7247). Available from: https://doi.org/10.1038/nature08026
11. Peng W, Santos MD, Li Y, Tomchick DR, Orth K. High-resolution cryo-EM structures of the <I>E. coli</I> hemolysin ClyA oligomers. PLOS ONE. 2019; 14 (5). Available from: https://doi.org/10.1371/journal.pone.0213423
12. Wilson JS, Churchill-Angus AM, Davies SP, Sedelnikova SE, Tzokov, SB, Rafferty JB, <I>et al</I>. Identification and structural analysis of the tripartite α-pore forming toxin of <I>Aeromonas hydrophila</I>. Nature Communications. 2019; 10 (1). Available from: https://doi.org/10.1038/s41467-019-10777-x
13. Brauning B, Groll M. Structural and Mechanistic Features of ClyA-Like α-Pore-Forming Toxins. Toxins. 2018; 10 (9). Available from: https://doi.org/10.3390/toxins10090343
14. Wai SN, Lindmark B, Söderblom T, Takade A, Westermark M, Oscarsson J, <I>et al</I>. Vesicle-Mediated Export and Assembly of Pore-Forming Oligomers of the Enterobacterial ClyA Cytotoxin. Cell. 2003; 115 (1). Available from: https://doi.org/10.1016/s0092-8674(03)00754-2
15. Rudkin JK, McLoughlin RM, Preston A, Massey RC. Bacterial toxins: Offensive, defensive, or something else altogether?. PLOS Pathogens. 2017; 13 (9). Available from: https://doi.org/10.1371/journal.ppat.1006452
16. Crnković A, Srnko M, Anderluh G. Biological Nanopores: Engineering on Demand. Life. 2021; 11 (1). Available from: https://doi.org/10.3390/life11010027
17. Murase K. Cytolysin A (ClyA): A Bacterial Virulence Factor with Potential Applications in Nanopore Technology, Vaccine Development, and Tumor Therapy. Toxins. 2022; 14 (2). Available from: https://doi.org/10.3390/toxins14020078
18. Oscarsson J, Mizunoe Y, Li L, Lai XH, Wieslander Å, Uhlin BE. Molecular analysis of the cytolytic protein ClyA (SheA) from <I>Escherichia coli</I>. Molecular Microbiology. 1999; 32 (6). Available from: https://doi.org/10.1046/j.1365-2958.1999.01435.x
19. Mishra S, Verma SS, Rai V, Awasthee N, Arya JS, Maiti KK, Gupta SC, <I>et al</I>. <I>Curcuma raktakanda</I> induces apoptosis and suppresses migration in cancer cells: Role of reactive oxygen species. Biomolecules. 2019; 9 (4). Available from: https://doi.org/10.3390/biom9040159
20. Alouf JE, Ladant D, Popoff MR. The comprehensive sourcebook of bacterial protein toxins. Elsevier. 2005; Available from: https://doi.org/10.1016/B978-0-12-088445-2.X5000-8
21. Mousavi SM, Archangi B, Zolgharnein H, Zamani I. Biocolorant “prodigiosin” interferes with the growth of biofouling bacteria: an in vitro and in silico approach. Pigment & Resin Technology. 2022; 51 (1). Available from: https://doi.org/10.1108/prt-07-2020-0079
22. Mousavi S, Archangi B, Zamani I. Antibacterial properties of bacteriocin purified from <I>Serratia marcescens</I> and computerized assessment of its interaction with antigen 43 in <I>Escherichia coli</I>. Archives of Razi Institute. 2023; 78 Available from: https://doi.org/10.32592/ari.2023.78.6.1738
23. Zahaf N-I, Schmidt G. Bacterial Toxins for Cancer Therapy. Toxins. 2017; 9 (8). Available from: https://doi.org/10.3390/toxins9080236
24. Sedighi M, Zahedi Bialvaei A, Hamblin MR, Ohadi E, Asadi A, Halajzadeh M, <I>et al</I>. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Medicine. 2019; 8 (6). Available from: https://doi.org/10.1002/cam4.2148
25. Ryan RM, Green J, Williams PJ, Tazzyman S, Hunt S, Harmey JH, <I>et al</I>. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Therapy. 2009; 16 (3). Available from: https://doi.org/10.1038/gt.2008.188
26. Li Y, Zhao R, Cheng K, Zhang K, Wang Y, Zhang Y, <I>et al</I>. Bacterial outer membrane vesicles presenting programmed death 1 for improved cancer immunotherapy via immune activation and checkpoint inhibition. ACS nano. 2020; 14 Available from: https://doi.org/10.1021/acsnano.0c03776
27. Brune KD, Leneghan DB, Brian IJ, Ishizuka AS, Bachmann MF, Draper SJ, <I>et al</I>. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Scientific Reports. 2016; 6 (1). Available from: https://doi.org/10.1038/srep19234
28. Cheng K, Zhao R, Li Y, Qi Y, Wang Y, Zhang Y, <I>et al</I>. Bioengineered bacteria-derived outer membrane vesicles as a versatile antigen display platform for tumor vaccination via Plug-and-Display technology. Nature Communications. 2021; 12 (1). Available from: https://doi.org/10.1038/s41467-021-22308-8
29. Gujrati V, Kim S, Kim S-H, Min JJ, Choy HE, Kim SC, <I>et al</I>. Bioengineered Bacterial Outer Membrane Vesicles as Cell-Specific Drug-Delivery Vehicles for Cancer Therapy. ACS Nano. 2014; 8 (2). Available from: https://doi.org/10.1021/nn405724x
30. Huang W, Wang S, Yao Y, Xia Y, Yang X, Li X, <I>et al</I>. Employing <I>Escherichia coli</I>-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against <I>Acinetobacter baumannii</I> infection. Scientific Reports. 2016; 6 (1). Available from: https://doi.org/10.1038/srep37242
31. Yang Z, Hua L, Yang M, Liu SQ, Shen J, Li W, <I>et al</I>. RBD-modified bacterial vesicles elicited potential protective immunity against SARS-CoV-2. Nano Letters. 2021; 21 Available from: https://doi.org/10.1021/acs.nanolett.1c00680
32. Thomas SC, Madaan T, Kamble NS, Siddiqui NA, Pauletti GM, Kotagiri N. Engineered Bacteria Enhance Immunotherapy and Targeted Therapy through Stromal Remodeling of Tumors. Advanced Healthcare Materials. 2022; 11 (2). Available from: https://doi.org/10.1002/adhm.202101487
33. Rho HW, Choi MJ, Lee JN, Park JW, Kim JS, Park BH, <I>et al</I>. Cytotoxic mechanism of <I>Vibrio vulnificus</I> cytolysin in CPAE cells. Life Sciences. 2002; 70 (16). Available from: https://doi.org/10.1016/s0024-3205(02)01480-7
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Published By Sri Devaraj Urs Academy of Higher Education, Kolar, Karnataka
Subscribe now for latest articles and news.